doi: https://doi.org/10.1101/2023.05.31.542821
Abstract
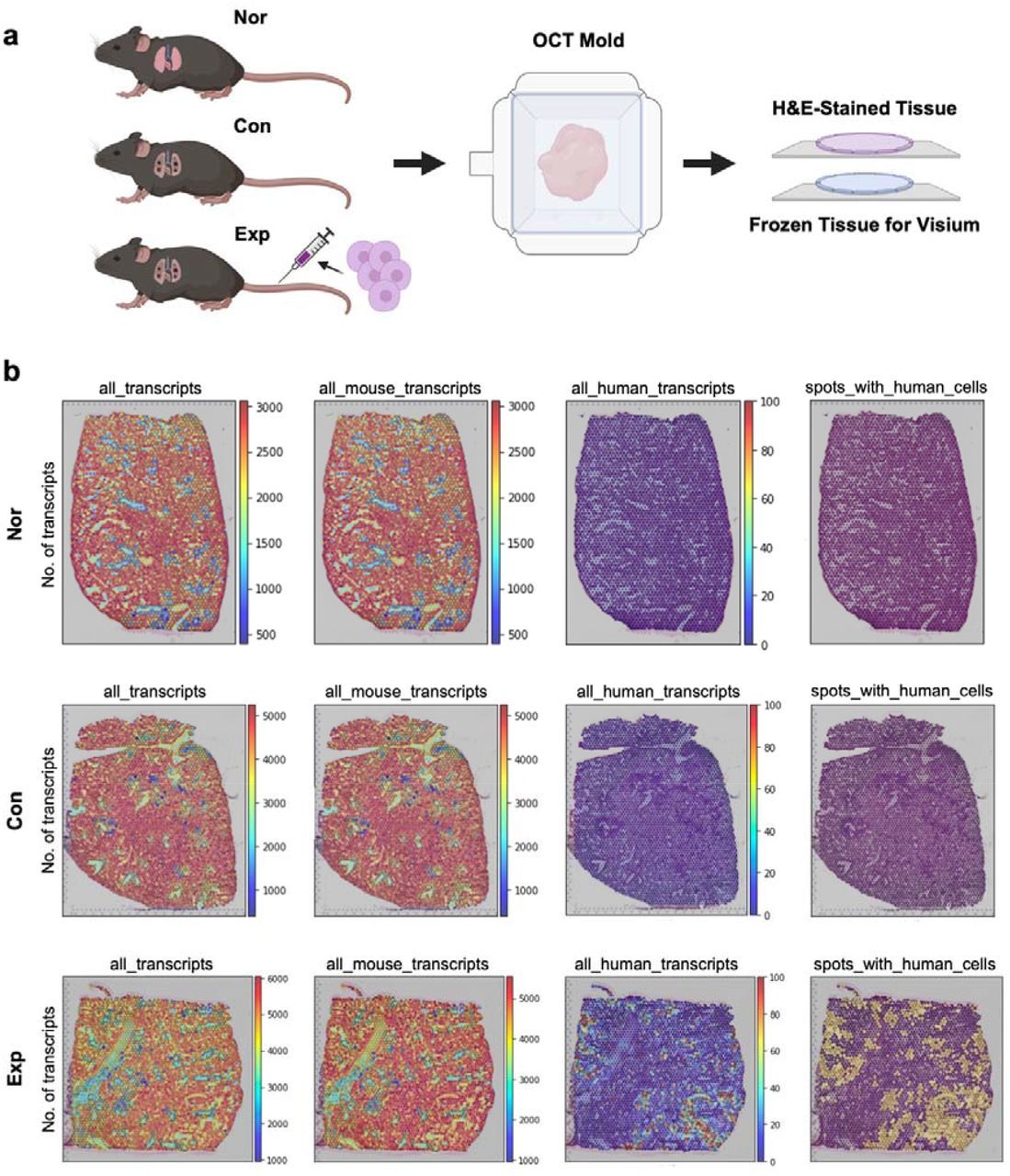
Recently, cell therapy has emerged as a promising treatment option for various disorders. Given the intricate mechanisms of action (MOA) and heterogenous distribution in target tissues inherent to cell therapy, it is necessary to develop more sophisticated, unbiased approaches to evaluate the distribution of administered cells and the molecular changes at a microscopic level. In this study, we present a label-free approach for assessing the tissue distribution of administered human mesenchymal stem cells (hMSCs) and their MOA, leveraging spatially resolved transcriptomics (ST) analysis. We administered hMSCs to mouse model of lung fibrosis and utilized ST to visualize the spatial distribution of hMSCs within the tissue. This was achieved by capitalizing on interspecies transcript differences between human and mouse. Furthermore, we could examine molecular changes associated with the spatial distribution of hMSCs. We suggest that our method has the potential to serve as an effective tool for various cell-based therapeutic agents.
Authors
Park J, Lee D, Lee J, Lee D, Song I, Park H, Choi H, Im H.(2023)
