doi: https://doi.org/10.1101/2023.02.07.527488
Abstract
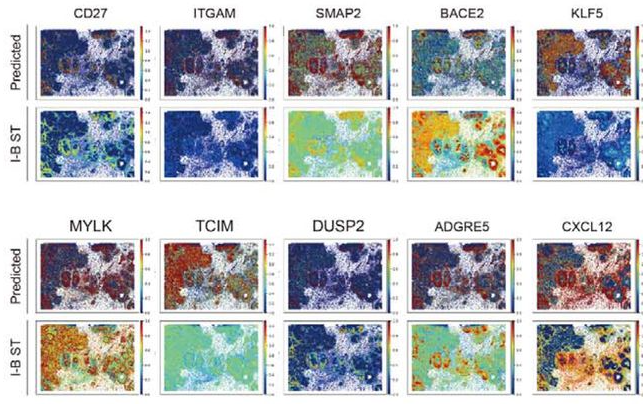
Spatial transcriptomics (ST) technologies provide comprehensive biological insights regarding cell-cell interactions and peri-cellular microenvironments. ST technologies are divided into two categories: imaging-based (I-B) and barcode-based (B-B). I-B ST technologies provide high resolution and sensitivity but have limited gene coverage. B-B ST technologies can analyze the whole transcriptome but have lower spatial resolution. To address these limitations, we propose a deep learning-based model that integrates I-B and B-B ST technologies to increase gene coverage while preserving high resolution. A model, trained by a neural network with an adversarial loss based on I-B and B-B datasets from human breast cancer tissue, was able to extend gene coverage to whole transcripts-level and accurately predict gene expression patterns in the I-B dataset with a high resolution. This novel methodology, named GeneDART, could enable researchers to utilize B-B and I-B ST datasets in a complementary way.
Authors
Ohn J, Lee D, Choi H.(2023)
